Comparative Glycomics Analyses of Three Functionally Distinct Secretions of the Garden Snail

What Has Been Achieved:
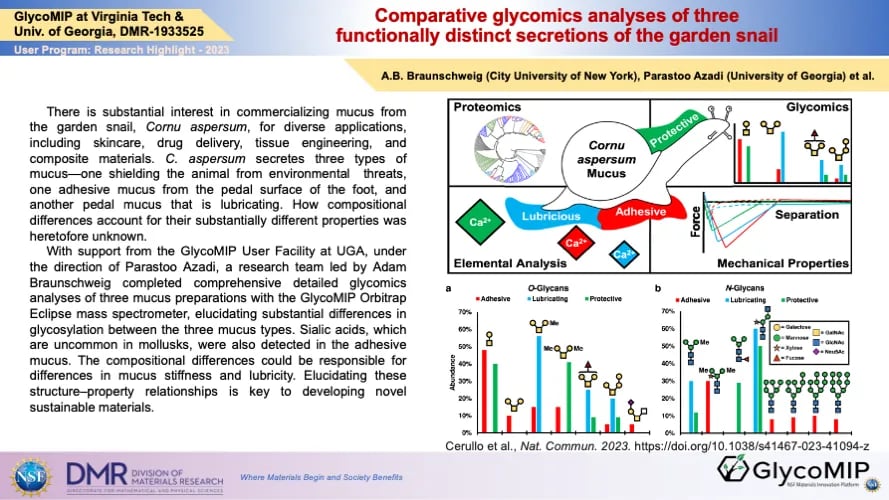
O- and N-linked glycans were released from three types of snail mucus and analyzed by two different mass spectrometry methods, yielding comprehensive detailed glycomics information for each mucus type. Significant differences and unique glycan patterns were discovered in the different mucus types, consistent with their functional differences.
Importance of the Achievement:
Glycomaterials are abundant in nature and take on many forms, ranging from crystalline fibrils to water-soluble molecules. The structural characteristics that govern their natural functions and properties are not well understood. Published in Nature Communications, this user-led study advances our understanding of the structure–property relationships of natural glycomaterials and is a step forward toward the rational design of novel glycomaterials with desired properties and diverse potential applications. The project was reported on by multiple outlets, including NPR.
Unique Feature(s) of the MIP that Enabled this Achievement:
The achievement was enabled by the preparative and analytical expertise of GlycoMIP scientists at UGA as well as the GlycoMIP-funded Orbitrap Eclipse mass spectrometry instrument.
Publication:
Cerullo, A.R.; McDermott, M.B.; Pepi, L.E.; Liu, Z. -L.; Barry, D.; Zhang, S.; Yang, X.; Chen, X.; Azadi, P.; Holford, M.; Braunschweig, A.B. Comparative mucomic analysis of three functionally distinct cornu aspersum secretions. Nature Communications 2023, 14, 5361. https://doi.org/10.1038/s41467-023-41094-z.
Acknowledgement statement:
This work is supported by a CUNY Science Scholarship and a CUNY Llewellyn Fellowship (A.R.C.); the Air Force Office of Scientific Research grants FA9550-19-1-0220, FA9550-18-1-0142, FA9550-23-1-0230 and the National Science Foundation grant DMR-2212139 (A.B.B.); the Complex Carbohydrate Research Center (Athens, GA), the National Institutes of Health-funded R24 grant NIH-R24GM137782 and NSF GlycoMIP, a National Science Foundation Materials Innovation Platform funded through Cooperative Agreement DMR-1933525 (L.E.P., X.Y., and P.A.); the Office of Naval Research N00014-18-1-2492 (X.C.); the Army Edu- cational Outreach Program (Rochester, NY) and Harlem Educational Activities Fund (New York, NY) (D.B. and A.B.B.). the Allen Institute’s Distinguished Investigator Award and NIH SPEECH Pilot Project U54CA221704, U54CA221705 (M.H.). Taylor Knapp and Peconic Escargot (Cutchogue, NY) are acknowledged for providing the animals used in this study. Genevieve Arroyo, Nicholas Mueller, and Robert Gullery are acknowledged for assisting in the collection of snail mucus. The Gard- ner, Cassacia, and Ulijn labs at the CUNY Advanced Science Research Center are acknowledged for allowing use of lab equipment and space to conduct experiments. NYU’s Genome Technology Center is acknowledged for conducting the transcriptomic sequencing with support from the Laura and Issac Perlmutter Cancer Center (Cancer Center Support Grant P30CA016087). The Clinical Proteomics Platform at the RIMUHC, McGill is acknowledged for conducting the proteomics experiments and analysis.